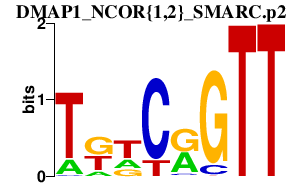
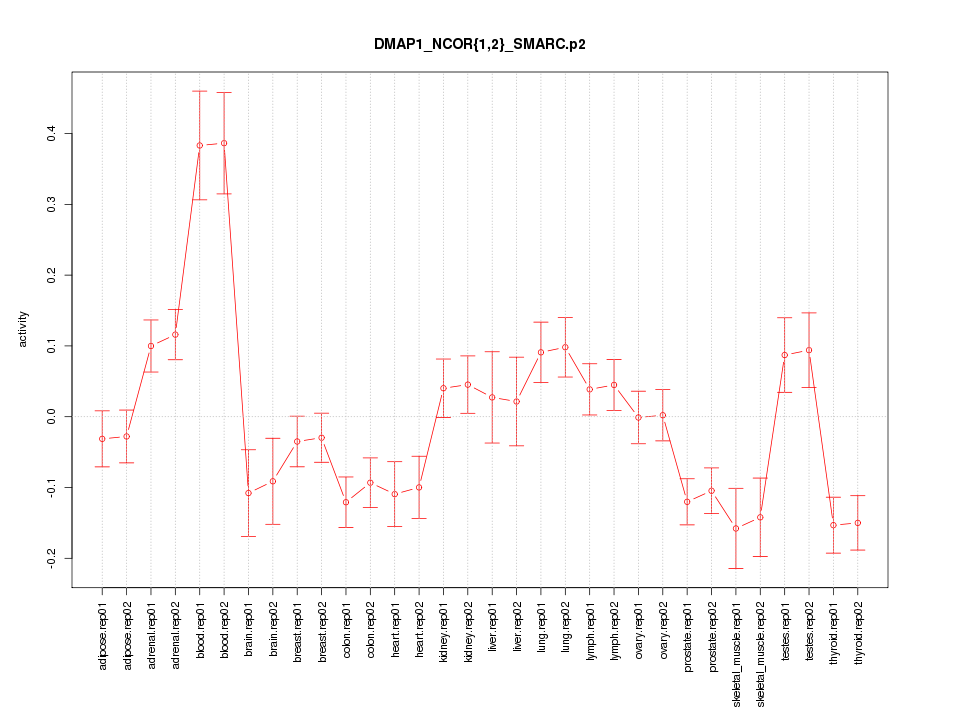
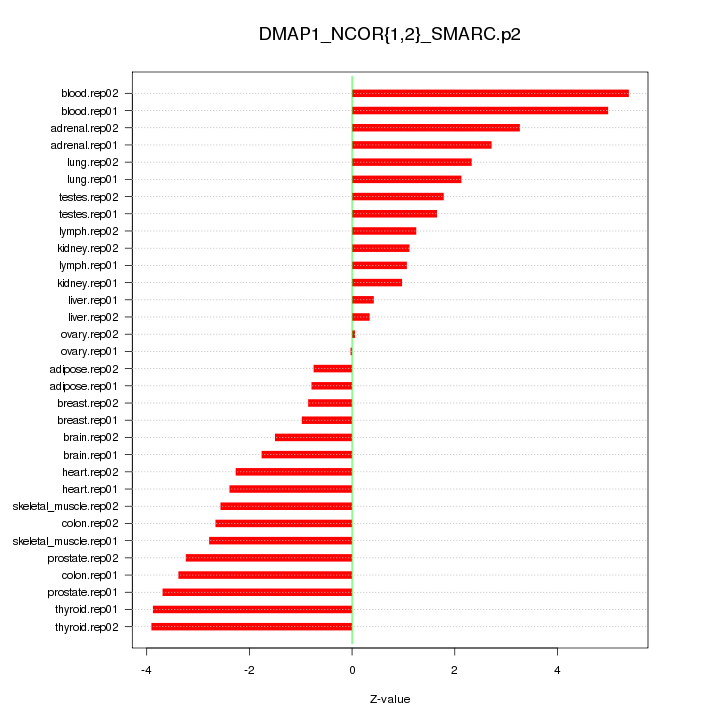
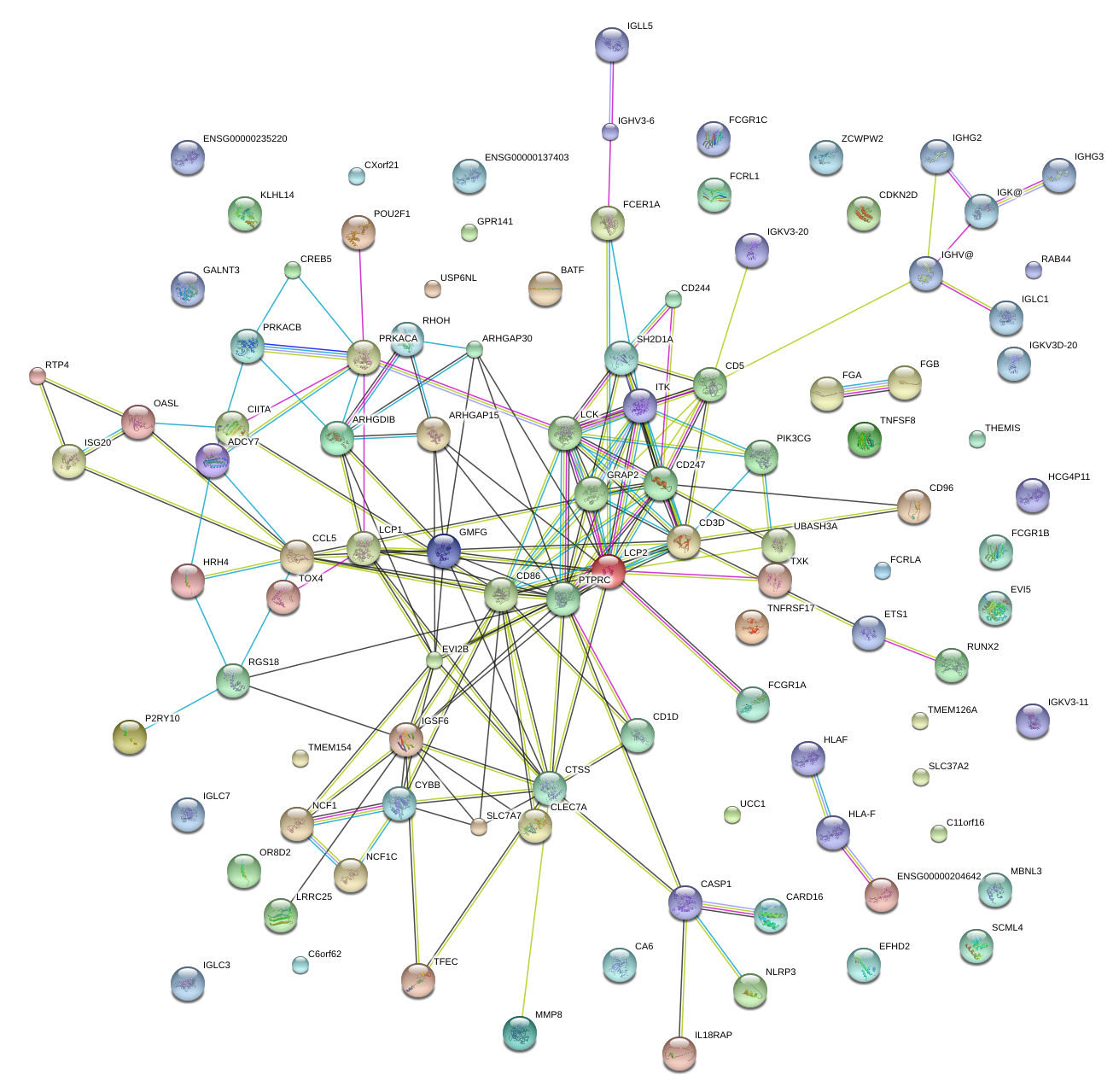
|
chrX_+_78200828
|
11.227
|
NM_014499
NM_198333
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10
|
|
chr7_-_115670828
|
10.795
|
NM_001018058
NM_012252
|
TFEC
|
transcription factor EC
|
|
chr6_-_108145507
|
9.890
|
NM_198081
|
SCML4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chr11_+_60869929
|
9.792
|
NM_014207
|
CD5
|
CD5 molecule
|
|
chrX_+_123480131
|
9.458
|
NM_001114937
NM_002351
|
SH2D1A
|
SH2 domain containing 1A
|
|
chr1_+_198608216
|
8.572
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr7_-_115670774
|
8.489
|
|
TFEC
|
transcription factor EC
|
|
chrX_-_131573638
|
8.447
|
NM_018388
NM_133486
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr1_-_150738232
|
8.440
|
|
CTSS
|
cathepsin S
|
|
chrX_+_37639246
|
8.307
|
NM_000397
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr1_+_32739711
|
8.255
|
NM_001042771
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr7_+_106505876
|
8.108
|
NM_002649
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr1_-_167487701
|
7.715
|
NM_000734
NM_198053
|
CD247
|
CD247 molecule
|
|
chr17_-_29641087
|
7.686
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr7_-_115670811
|
7.580
|
|
TFEC
|
transcription factor EC
|
|
chrX_+_37639306
|
7.564
|
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr11_-_118213321
|
7.509
|
NM_000732
NM_001040651
|
CD3D
|
CD3d molecule, delta (CD3-TCR complex)
|
|
chr17_-_34207308
|
7.365
|
NM_002985
|
CCL5
|
chemokine (C-C motif) ligand 5
|
|
chr7_+_74188308
|
7.338
|
NM_000265
|
NCF1
NCF1B
|
neutrophil cytosolic factor 1
neutrophil cytosolic factor 1B pseudogene
|
|
chr22_+_40322609
|
7.294
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr4_-_48136245
|
7.199
|
NM_003328
|
TXK
|
TXK tyrosine kinase
|
|
chr1_+_167298280
|
7.036
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chrX_-_30595883
|
6.949
|
NM_025159
|
CXorf21
|
chromosome X open reading frame 21
|
|
chr2_+_143886898
|
6.876
|
NM_018460
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr5_-_169724734
|
6.813
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr5_+_156607847
|
6.744
|
NM_005546
|
ITK
|
IL2-inducible T-cell kinase
|
|
chr1_-_150738292
|
6.688
|
NM_001199739
NM_004079
|
CTSS
|
cathepsin S
|
|
chr9_-_117692874
|
6.588
|
NM_001244
NM_001252290
|
TNFSF8
|
tumor necrosis factor (ligand) superfamily, member 8
|
|
chr1_-_150738255
|
6.541
|
|
CTSS
|
cathepsin S
|
|
chr5_-_138725544
|
6.499
|
NM_016459
|
MZB1
|
marginal zone B and B1 cell-specific protein
|
|
chr11_-_104905849
|
6.439
|
|
CASP1
|
caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase)
|
|
chr2_+_143886969
|
6.415
|
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr12_-_10282680
|
6.379
|
NM_022570
NM_197947
NM_197948
NM_197949
NM_197950
NM_197954
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chr9_+_127420690
|
6.273
|
|
MIR181A2HG
|
MIR181A2 host gene (non-protein coding)
|
|
chr5_-_169724817
|
6.083
|
NM_005565
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr5_-_169724782
|
6.050
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr16_-_21663962
|
6.048
|
NM_005849
|
IGSF6
|
immunoglobulin superfamily, member 6
|
|
chr1_+_198608136
|
5.870
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr16_-_21663908
|
5.686
|
|
IGSF6
|
immunoglobulin superfamily, member 6
|
|
chr5_-_169724614
|
5.589
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr11_-_102595623
|
5.361
|
NM_002424
|
MMP8
|
matrix metallopeptidase 8 (neutrophil collagenase)
|
|
chr14_-_106725232
|
5.329
|
|
ZCWPW2
IGHV3-23
IGKV3-20
IGK@
|
zinc finger, CW type with PWWP domain 2
immunoglobulin heavy variable 3-23
immunoglobulin kappa variable 3-20
immunoglobulin kappa locus
|
|
chr1_-_157789832
|
5.237
|
NM_001159397
NM_001159398
NM_052938
|
FCRL1
|
Fc receptor-like 1
|
|
chr3_+_111260925
|
5.198
|
NM_005816
NM_198196
|
CD96
|
CD96 molecule
|
|
chr4_+_40198526
|
5.084
|
NM_004310
|
RHOH
|
ras homolog gene family, member H
|
|
chr1_+_159259503
|
5.071
|
NM_002001
|
FCER1A
|
Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide
|
|
chr1_+_32716839
|
5.046
|
NM_005356
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr12_-_121476770
|
5.041
|
NM_003733
NM_198213
|
OASL
|
2'-5'-oligoadenylate synthetase-like
|
|
chr11_-_104972147
|
5.037
|
NM_001007232
NM_001223
NM_033292
NM_033293
NM_033294
NM_033295
|
CARD17
CASP1
|
caspase recruitment domain family, member 17
caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase)
|
|
chr1_+_192127591
|
4.939
|
NM_130782
|
RGS18
|
regulator of G-protein signaling 18
|
|
chr16_+_10971038
|
4.911
|
NM_000246
|
CIITA
|
class II, major histocompatibility complex, transactivator
|
|
chr11_-_104916041
|
4.866
|
NM_001017534
NM_052889
|
CARD16
|
caspase recruitment domain family, member 16
|
|
chr1_+_158149736
|
4.855
|
NM_001766
|
CD1D
|
CD1d molecule
|
|
chr6_+_36665627
|
4.774
|
|
RAB44
|
RAB44, member RAS oncogene family
|
|
chr19_-_39826625
|
4.745
|
|
GMFG
|
glia maturation factor, gamma
|
|
chr12_-_15114495
|
4.735
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr1_+_167298382
|
4.678
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr4_+_155484131
|
4.626
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr19_+_17516502
|
4.615
|
|
LOC100507535
|
uncharacterized LOC100507535
|
|
chr22_+_40297085
|
4.590
|
NM_004810
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr1_-_160832466
|
4.487
|
NM_001166663
NM_001166664
NM_016382
|
CD244
|
CD244 molecule, natural killer cell receptor 2B4
|
|
chr19_-_18508407
|
4.445
|
NM_145256
|
LRRC25
|
leucine rich repeat containing 25
|
|
chr3_+_121796695
|
4.437
|
NM_006889
NM_176892
|
CD86
|
CD86 molecule
|
|
chr11_-_8954534
|
4.396
|
NM_020643
|
C11orf16
|
chromosome 11 open reading frame 16
|
|
chr1_+_247579457
|
4.358
|
NM_001079821
NM_001127461
|
NLRP3
|
NLR family, pyrin domain containing 3
|
|
chr1_+_84630575
|
4.356
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr1_+_149754245
|
4.328
|
NM_000566
|
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64)
|
|
chr1_-_93257950
|
4.319
|
NM_005665
|
EVI5
|
ecotropic viral integration site 5
|
|
chr11_-_128392061
|
4.283
|
NM_001162422
NM_005238
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr2_+_103035148
|
4.230
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr19_-_10679328
|
4.195
|
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4)
|
|
chr4_-_153601085
|
4.164
|
NM_152680
|
TMEM154
|
transmembrane protein 154
|
|
chr19_-_39826676
|
4.101
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr6_-_138189313
|
4.091
|
|
|
|
|
chr16_+_50308020
|
4.088
|
|
ADCY7
|
adenylate cyclase 7
|
|
chr17_-_29641068
|
4.077
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr1_-_161039745
|
4.012
|
NM_001025598
NM_181720
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr14_-_23288912
|
3.997
|
NM_001126106
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr14_+_75988765
|
3.957
|
NM_006399
|
BATF
|
basic leucine zipper transcription factor, ATF-like
|
|
chr18_+_22040592
|
3.893
|
NM_001143828
NM_001160166
NM_021624
|
HRH4
|
histamine receptor H4
|
|
chr15_+_89182038
|
3.889
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr7_-_96633608
|
3.830
|
|
DLX6-AS1
|
DLX6 antisense RNA 1 (non-protein coding)
|
|
chr1_-_161039703
|
3.821
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr14_+_75988838
|
3.734
|
|
BATF
|
basic leucine zipper transcription factor, ATF-like
|
|
chr1_+_15736387
|
3.724
|
NM_024329
|
EFHD2
|
EF-hand domain family, member D2
|
|
chr6_+_29691116
|
3.703
|
NM_001098478
NM_001098479
NM_018950
|
HLA-F
|
major histocompatibility complex, class I, F
|
|
chr13_-_46756326
|
3.685
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr17_-_29641094
|
3.673
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr6_+_45296053
|
3.611
|
NM_001015051
NM_001024630
|
RUNX2
|
runt-related transcription factor 2
|
|
chr11_-_128391888
|
3.611
|
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr2_-_166650546
|
3.582
|
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr1_+_15736422
|
3.575
|
|
EFHD2
|
EF-hand domain family, member D2
|
|
chr7_+_37779995
|
3.520
|
NM_181791
|
GPR141
|
G protein-coupled receptor 141
|
|
chr6_-_24719286
|
3.520
|
NM_030939
|
C6orf62
|
chromosome 6 open reading frame 62
|
|
chr22_+_23229959
|
3.501
|
NM_001178126
|
IGLL5
IGLC1
|
immunoglobulin lambda-like polypeptide 5
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr4_-_155511838
|
3.497
|
NM_000508
NM_021871
|
FGA
|
fibrinogen alpha chain
|
|
chr1_+_9005921
|
3.479
|
NM_001215
|
CA6
|
carbonic anhydrase VI
|
|
chr10_-_11574273
|
3.472
|
NM_001080491
|
USP6NL
|
USP6 N-terminal like
|
|
chr16_+_12058963
|
3.464
|
NM_001192
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17
|
|
chr6_-_128222175
|
3.423
|
NM_001010923
NM_001164685
|
THEMIS
|
thymocyte selection associated
|
|
chr18_-_30352841
|
3.415
|
NM_020805
|
KLHL14
|
kelch-like 14 (Drosophila)
|
|
chr3_+_187086167
|
3.387
|
NM_022147
|
RTP4
|
receptor (chemosensory) transporter protein 4
|
|
chr11_+_124933197
|
3.349
|
|
SLC37A2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2
|
|
chr21_+_43823970
|
3.322
|
NM_001001895
NM_001243467
NM_018961
|
UBASH3A
|
ubiquitin associated and SH3 domain containing A
|
|
chr7_+_28452143
|
3.307
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr14_-_107199126
|
3.294
|
|
IGHG3
|
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr13_-_74707913
|
3.249
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr11_+_124933007
|
3.236
|
NM_001145290
NM_198277
|
SLC37A2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2
|
|
chr14_-_25103365
|
3.228
|
NM_004131
|
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1)
|
|
chr4_+_36285643
|
3.181
|
NM_001170700
|
DTHD1
|
death domain containing 1
|
|
chr13_+_108921976
|
3.166
|
NM_001145645
NM_006573
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chr2_-_166650770
|
3.143
|
NM_004482
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr18_+_72922725
|
3.135
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr16_-_28518131
|
3.112
|
NM_145659
|
IL27
|
interleukin 27
|
|
chr7_-_76829109
|
3.052
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chr1_-_47695442
|
3.048
|
NM_003189
|
TAL1
|
T-cell acute lymphocytic leukemia 1
|
|
chr2_-_166650705
|
3.046
|
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr3_-_157221127
|
3.042
|
NM_001167911
NM_001167912
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr1_+_40862471
|
3.042
|
NM_001198980
|
SMAP2
|
small ArfGAP2
|
|
chr1_+_84630374
|
3.041
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr12_-_53601051
|
3.032
|
|
ITGB7
|
integrin, beta 7
|
|
chr1_+_84609987
|
3.018
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr22_+_40297183
|
3.010
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr13_-_103719195
|
2.993
|
NM_000452
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 2
|
|
chrX_+_12924738
|
2.947
|
NM_138636
|
TLR8
|
toll-like receptor 8
|
|
chr1_+_198608244
|
2.945
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr17_+_6659356
|
2.942
|
|
XAF1
|
XIAP associated factor 1
|
|
chr3_-_3152057
|
2.936
|
NM_000564
NM_001243099
NM_175724
NM_175725
NM_175726
NM_175727
NM_175728
|
IL5RA
|
interleukin 5 receptor, alpha
|
|
chr2_-_161130151
|
2.925
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr10_-_14816895
|
2.923
|
NM_031453
|
FAM107B
|
family with sequence similarity 107, member B
|
|
chr16_+_57023411
|
2.917
|
|
NLRC5
|
NLR family, CARD domain containing 5
|
|
chr22_-_23922376
|
2.905
|
NM_020070
NM_152855
|
IGLL1
|
immunoglobulin lambda-like polypeptide 1
|
|
chr1_+_113162449
|
2.780
|
|
CAPZA1
|
capping protein (actin filament) muscle Z-line, alpha 1
|
|
chr21_-_34185943
|
2.729
|
NM_001162495
NM_001162496
NM_019596
|
C21orf62
|
chromosome 21 open reading frame 62
|
|
chr1_+_161676761
|
2.716
|
NM_001184866
NM_001184867
NM_001184870
NM_001184871
NM_001184872
NM_001184873
NM_032738
|
FCRLA
|
Fc receptor-like A
|
|
chr2_-_160472960
|
2.711
|
NM_013450
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr12_+_25205180
|
2.707
|
NM_001204126
NM_001204127
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr16_-_11350038
|
2.688
|
NM_003745
|
SOCS1
|
suppressor of cytokine signaling 1
|
|
chr6_+_31539875
|
2.687
|
NM_001159740
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr14_-_106691698
|
2.681
|
|
|
|
|
chr1_-_200377262
|
2.660
|
|
ZNF281
|
zinc finger protein 281
|
|
chr1_+_113162074
|
2.635
|
NM_006135
|
CAPZA1
|
capping protein (actin filament) muscle Z-line, alpha 1
|
|
chrX_-_49121171
|
2.608
|
NM_001114377
NM_014009
|
FOXP3
|
forkhead box P3
|
|
chr1_+_113162421
|
2.567
|
|
CAPZA1
|
capping protein (actin filament) muscle Z-line, alpha 1
|
|
chr1_-_207095198
|
2.567
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr17_-_34417386
|
2.555
|
NM_002983
|
CCL3
|
chemokine (C-C motif) ligand 3
|
|
chr12_-_9885859
|
2.517
|
NM_001253748
NM_001253750
NM_172004
|
CLECL1
|
C-type lectin-like 1
|
|
chr1_+_158259558
|
2.505
|
NM_001765
|
CD1C
|
CD1c molecule
|
|
chr5_-_176738831
|
2.490
|
|
MXD3
|
MAX dimerization protein 3
|
|
chr1_+_113162456
|
2.489
|
|
CAPZA1
|
capping protein (actin filament) muscle Z-line, alpha 1
|
|
chr1_+_84630014
|
2.485
|
NM_001242857
NM_001242858
NM_001242859
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr3_-_49851217
|
2.471
|
|
UBA7
|
ubiquitin-like modifier activating enzyme 7
|
|
chr4_-_40516567
|
2.465
|
|
RBM47
|
RNA binding motif protein 47
|
|
chr17_+_6659155
|
2.455
|
NM_017523
NM_199139
|
XAF1
|
XIAP associated factor 1
|
|
chr1_+_48688356
|
2.450
|
NM_001011547
NM_001135181
|
SLC5A9
|
solute carrier family 5 (sodium/glucose cotransporter), member 9
|
|
chr3_-_157221362
|
2.425
|
NM_001167915
NM_001167916
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr2_+_113885137
|
2.403
|
NM_173842
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr1_+_113217039
|
2.397
|
NM_001130079
NM_020963
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse)
|
|
chr5_-_41071404
|
2.322
|
NM_173489
|
HEATR7B2
|
HEAT repeat family member 7B2
|
|
chr19_-_10679620
|
2.313
|
NM_001800
NM_079421
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4)
|
|
chr15_-_45003513
|
2.310
|
|
|
|
|
chr7_-_92777657
|
2.283
|
NM_152703
|
SAMD9L
|
sterile alpha motif domain containing 9-like
|
|
chr3_-_195938256
|
2.269
|
NM_001039617
|
ZDHHC19
|
zinc finger, DHHC-type containing 19
|
|
chr4_-_122085475
|
2.268
|
NM_024873
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr17_-_80407601
|
2.267
|
NM_001193655
|
C17orf62
|
chromosome 17 open reading frame 62
|
|
chr1_+_948846
|
2.266
|
NM_005101
|
ISG15
|
ISG15 ubiquitin-like modifier
|
|
chr3_+_63953419
|
2.261
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr12_+_11802905
|
2.257
|
|
ETV6
|
ets variant 6
|
|
chr1_-_200377488
|
2.257
|
|
ZNF281
|
zinc finger protein 281
|
|
chr6_-_32908583
|
2.243
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr6_+_26365397
|
2.241
|
NM_001197247
NM_001197249
NM_007047
NM_001197248
|
BTN3A2
|
butyrophilin, subfamily 3, member A2
|
|
chr6_-_133035180
|
2.239
|
NM_004666
|
VNN1
|
vanin 1
|
|
chr6_-_149806116
|
2.235
|
NM_207360
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr12_+_11802726
|
2.233
|
NM_001987
|
ETV6
|
ets variant 6
|
|
chr15_+_80364902
|
2.218
|
NM_001242915
NM_001242916
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr17_+_7462064
|
2.215
|
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chr12_+_54447660
|
2.207
|
NM_153633
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr3_+_45927995
|
2.204
|
NM_006641
NM_031200
|
CCR9
|
chemokine (C-C motif) receptor 9
|
|
chr6_+_26402464
|
2.204
|
NM_001145008
NM_001145009
NM_007048
NM_194441
|
BTN3A1
|
butyrophilin, subfamily 3, member A1
|
|
chr3_-_112693711
|
2.192
|
NM_138806
NM_138939
NM_138940
NM_170780
|
CD200R1
|
CD200 receptor 1
|
|
chr6_+_105404922
|
2.147
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr15_+_45003659
|
2.147
|
NM_004048
|
B2M
|
beta-2-microglobulin
|
|
chr16_-_50715240
|
2.136
|
NM_001144972
NM_153337
NM_182854
|
SNX20
|
sorting nexin 20
|
|
chr5_-_149792276
|
2.129
|
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr11_+_5710926
|
2.122
|
|
TRIM22
|
tripartite motif containing 22
|
|
chr13_+_28712379
|
2.117
|
NM_175854
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr4_-_39523132
|
2.105
|
|
UGDH
|
UDP-glucose 6-dehydrogenase
|
|
chr13_-_26789062
|
2.103
|
|
RNF6
|
ring finger protein (C3H2C3 type) 6
|
|
chr17_-_47287935
|
2.083
|
NM_001198754
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2
|
|
chr12_-_53600999
|
2.077
|
NM_000889
|
ITGB7
|
integrin, beta 7
|
|
chr11_+_118754474
|
2.077
|
NM_001716
|
CXCR5
|
chemokine (C-X-C motif) receptor 5
|
|
chr5_-_149465989
|
2.038
|
|
CSF1R
|
colony stimulating factor 1 receptor
|
|
chr9_+_5890801
|
2.024
|
NM_005511
|
MLANA
|
melan-A
|
|
chr1_-_120935943
|
1.998
|
NM_001004340
NM_001017986
NM_001244910
|
FCGR1A
FCGR1B
|
Fc fragment of IgG, high affinity Ia, receptor (CD64)
Fc fragment of IgG, high affinity Ib, receptor (CD64)
|
|
chr12_-_49319211
|
1.998
|
NM_001143782
NM_016594
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chrX_-_19688991
|
1.975
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr3_-_167371288
|
1.973
|
NM_178824
|
WDR49
|
WD repeat domain 49
|
|
chr4_-_38806410
|
1.952
|
NM_003263
|
TLR1
|
toll-like receptor 1
|
|
chr2_+_182322265
|
1.948
|
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr1_-_169555624
|
1.939
|
|
F5
|
coagulation factor V (proaccelerin, labile factor)
|